This is my talk from the 2023 CEIRR annual meeting. You can find my slides here as well as embedded at the bottom of the page.
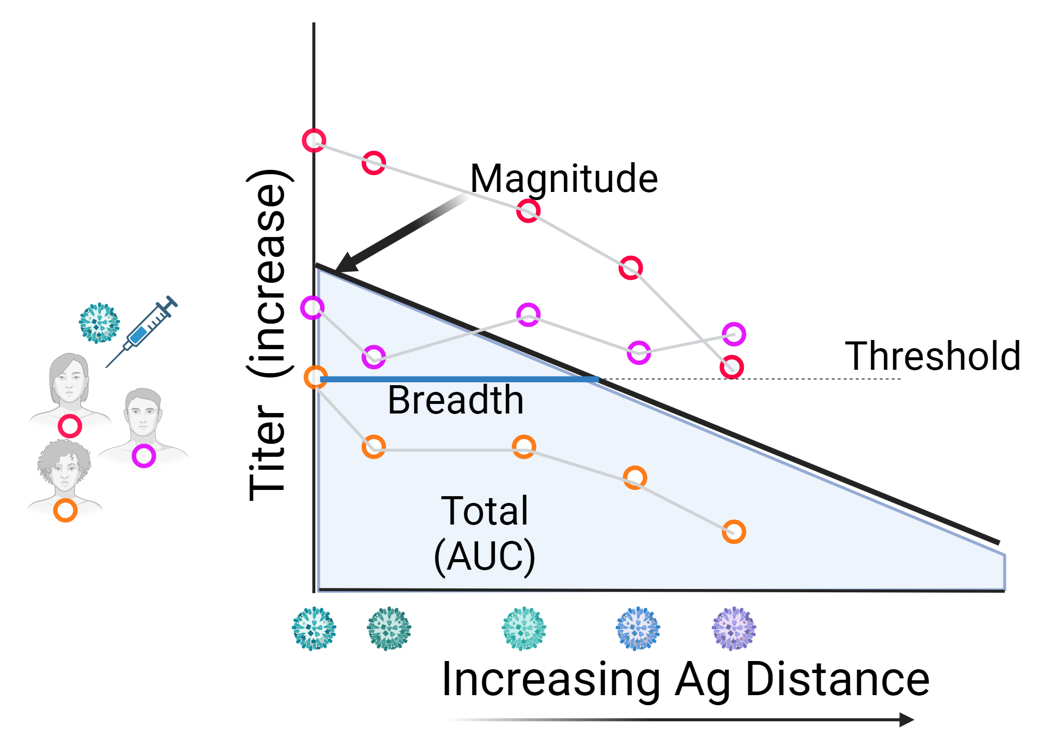
Summary: We’re currently working on how to quantify the “breadth” of a vaccine based on immunogenicity measurements from cohort studies. In our framework, we compute the antigenic distance of all the strains used for immune measurements and use the distance to creature antibody landscapes. From the antibody landscapes, we can compute summary statistics for the strength and breadth of the vaccine which we believe are more robust to the choice of strains used in the study than traditional methods. However, there are several methodological issues, primarily the issue of data points at the limit of detection, that we need to address to fairly test our framework. We are currently working on Bayesian hiearchical models as a strategy for mitigating the limitations of our method.
Official abstract: HAI titer is a commonly used metric for assessing the immunogenicity of the seasonal influenza vaccine to the virus strains used in the formulation of the vaccine antigen. A next-generation influenza vaccine should induce protection to similar strains, but an ideal vaccine candidate would also elicit a response to other variants. An antigenic distance measurement is a way to quantify how different two variants are and can be combined with immunogenicity measurements to a panel of different viral strains following vaccination in order to assess the breadth of the vaccine response. Building on previous literature on antibody landscapes, we propose a Bayesian hierarchical modeling framework for reducing noise across individual antibody landscapes and obtain measurements of the homologous and heterologous vaccine response that take antigenic distance into account. Various weighting systems can then be applied to our measurements in order to fairly judge vaccine candidates in different scenarios. We examine the strengths and limitations of our metrics by using both generative simulations and a case study with real-world immunogenicity data.